Generic open-access data repositories
Overview
Teaching: 45 min
Exercises: 0 minQuestions
What is an open-acess repository?
What is the difference between a generic and a domain-specific data repository?
What are examples of open-access repositories?
How can I upload my dataset to an open data repository?
Objectives
Understand where generic open-access repositories stand in the research data life cycle.
Be able to upload a dataset to one of the open-access repositories.
1. Table of contents
- 1. Introduction
- 3. Real-life example with Zenodo
- 4. FigShare
- 5. Dryad
- 4. Real-life exercise
- 5. Resources
1. Introduction
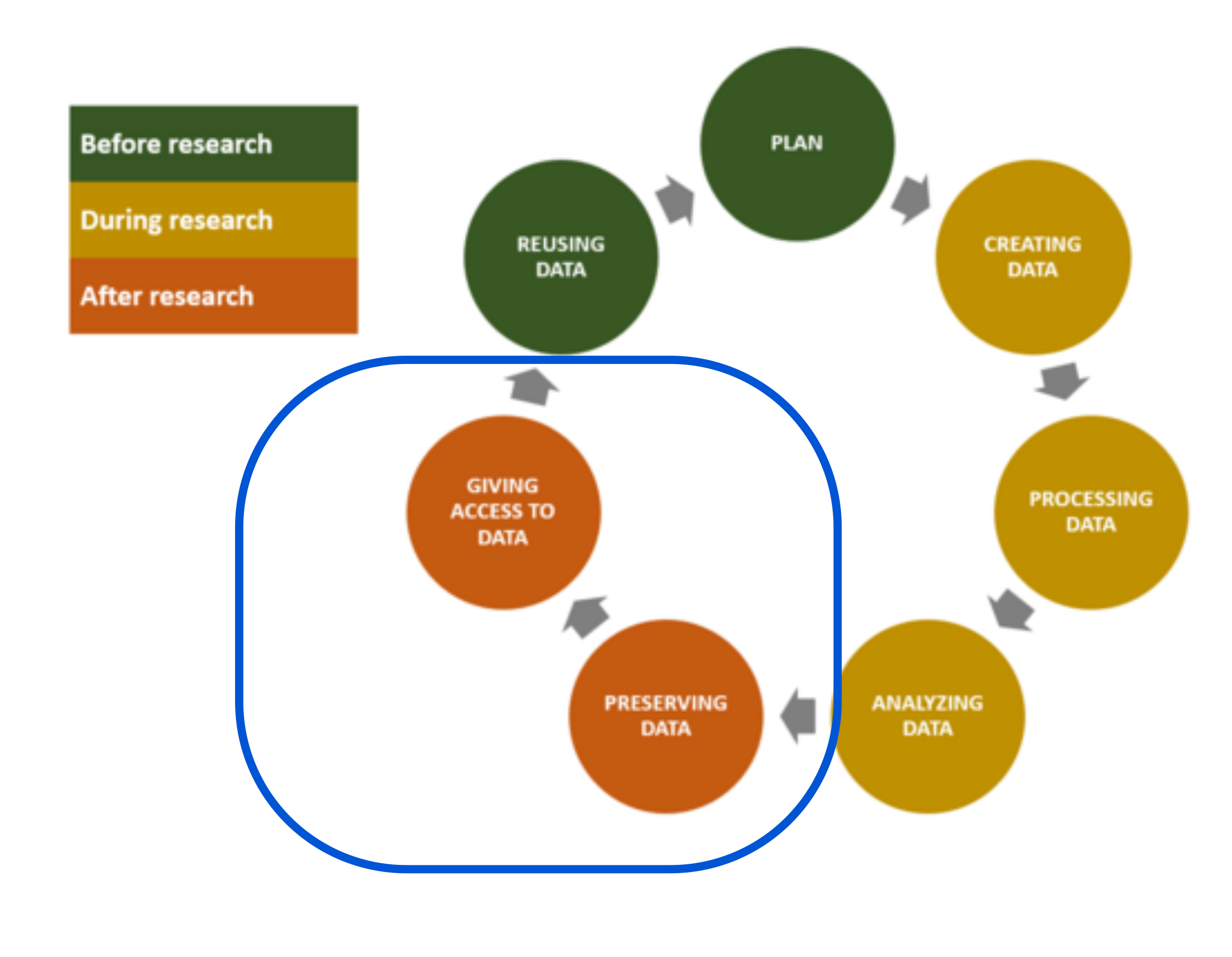
In this section, we will see how to preserve and support access to your research data generated during the course of your work. This episode on steps 5 and 6 of the Research Data Life Cycle.
Open Access often refers to free access to research publications. In this episode, Open Access will strictly refer to research outputs that are not publications themselves but rather additional resources collected that support the research conclusions and related publications.
1.1 Definition of an open-access repository
An open-access repository is a digital hosting service for the safe storage, data annotation and retrieval of research outputs. Open Access here means that datasets are made freely accessible either instantly or after an embargo period (becomes free after a certain time).
Question
Do you already know examples of open-access repositories?
Answer
If you continue the lesson, you will discover that Zenodo, FigShare or Dryad are examples of open-access repositories.
1.2 Open and public data
Open access does not necessary imply that all datasets are made public directly after upload to the data repository. Rather, these datasets should be made accessible and indicate on which conditions these can be used by others (i.e. license).
Taken from the EU FOSTER portal
Question
What would be a real-life example of a dataset that should be made accessible (open) but not public (keep private)?
Can you name a few examples?Answer
Clinical studies in which human patient data are collected is such an example of research data that should be accessible (by other clinical researchers for instance) but should not be made public. DNA sequencing is also treated
3. Real-life example with Zenodo
3.1 Short description of Zenodo
3.2 Upload a dataset through a web interface
In this section, you will mimick a real-life example. You have just obtained statistics on a rabbit population.
Download a test dataset. Description of the dataset
If not already done, create an account on Zenodo. If you have a GitHub or an ORCID account you can use them to login. Otherwise, you will have to enter an email address and a valid password.
Once logged in, you can then upload the test dataset by clicking on the “upload” button.
Sandbox: Zenodo has a “sandbox” website that is made to test data upload: access it here.
3.3 Programmatic upload using Python
While data upload using a web interface is convenient, it has a limited capacity whenever you have many files to upload. If your internet connection also gets interrupted, it could be that your data upload will get interrupted.
An alternative is to log in a server (which stays on) and transfer your data from that server to Zenodo using its API (application programming interface). Zenodo API provides a set of actions that a user can perform to modify and alter the datasets without having to understand the ins and outs of the Zenodo file management system.
- Post HTTP method.
- Requests Python package for HTTP interface.
- API ? + Location of the Zenodo API
- Access token generation
- ipython interface +
4. FigShare
5. Dryad
4. Real-life exercise
5. Resources
5.1 Open-access repositories
5.2 Open data
Key Points
Next-Generation Sequencing techniques are massively parallel cDNA sequencing.