Introduction to dataset #2 (methylation and age)
Overview
Teaching: 30 min
Exercises: 30 minQuestions
How are gene expression levels distributed within a RNA-seq experiment?
Objectives
Be able to calculate size factors and normalize counts using
DESeq2.
Table of contents
Introduction
Aging
Aging (also spelled ageing) or is the process of becoming older. From a biological perspective, the term “aging” is not clearly defined as it can refer to aging of human beings, animals or other organisms. In this context chronological age and biological age are frequently confused. An individual’s age is usually defined based on chronological time, but on the other hand it is well accepted that individuals age at different rates. More generally, aging can also refer to time-related changes in cells within an organism (cellular senescence) or to changes in the population of a species (population aging). In this practical, we will focus on the aging of human individuals.
In humans, aging represents the accumulation of changes in a human being over time, encompassing physical, psychological, and social change. At the cellular and molecular levels, the process of aging results in a wide range of changes, which include senescence, telomere shortening, and changes in gene expression.

Epigenetic clock
Epigenetic patterns also change over the lifespan, suggesting that epigenetic changes may constitute an important component of the aging process. The epigenetic mark that has mostly been studied is DNA methylation, the presence of methyl groups at CpG dinucleotides. These dinucleotides are often located near gene promoters and associate with gene expression levels. Thus, DNA methylation can play essential roles during development, acting through the regulation of gene expression.
The measure of age using DNA methylation levels can be called an epigenetic clock.
CpG islands
In genomes, some regions contain a high amount of CG sites (one cytosine followed by one guanine) and are called CpG islands. The cytosine of a CG site within a CpG island can be the target for 5-methylation which in turn can alter nearby gene expression.
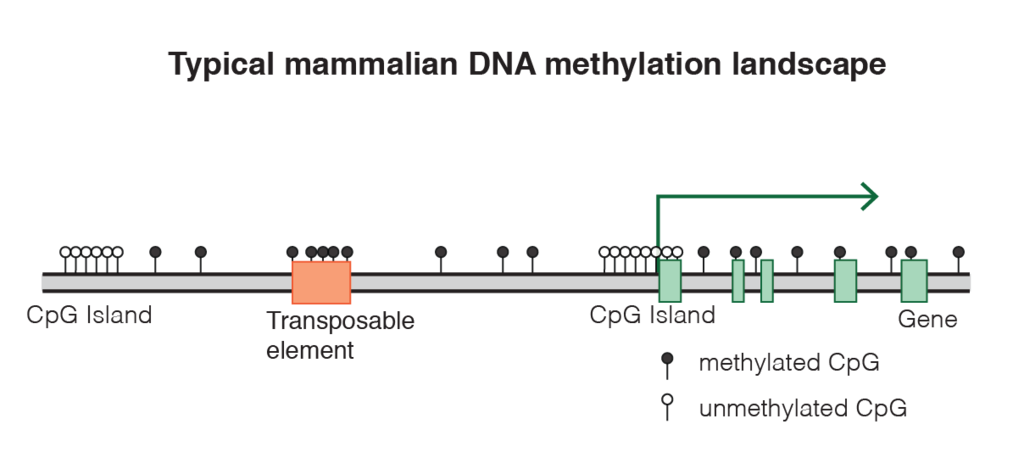
Approximately 60–70% of genes have a CpG island associated with their promoters, and promoters can be classified according to their CpG density. Levels of DNA methylation at a promoter-associated CpG island are generally negatively associated with gene expression, although some specific genes show the opposite effect. Interestingly, this negative correlation is not upheld when comparing expression and DNA methylation for a specific gene across individuals. Conversely, DNA methylation in the gene body is often positively associated with levels of gene expression. Although direct causal relationships between DNA methylation, gene expression and mechanisms of aging have not been established so far, it has been shown that systematic age-related changes in DNA methylation do occur.
Specific tools such as microarrays have been designed to measure the level of methylation of thousands of CpG islands simultaneously. For instance, the Illumina Methylation Assay can be used to measure simultaneously CpG dinucleotides from the Human genome. In total, 14,495 genes can have their CG site methylation state assayed.
Dataset description
Provenance
The dataset contains the genome-wide DNA methylation profiles of saliva samples from male identical twins from age 21 to 55.
The Illumina Infinium 27k Human DNA methylation Beadchip v1.2 was used to obtain DNA methylation profiles across approximately 27,000 CpGs in bisulfite converted DNA. Samples included 34 pairs of identical twins and one set of identical triplets, and 13 hybridisations were technical replicates of a selection of those samples.
Beta values
Beta values indicate the extent of DNA methylation at a given site as measured by the Values range from 0 (completely unmethylated CG site) to 1 (completely methylated).
Get started
Lesson section objective
In this practical you will perform genome-wide epigenetic analyses to investigate the relationship between human aging and DNA methylation.
Checklist
- Download the CpG methylation and age datasets: https://doi.org/10.5281/zenodo.4054842.
- Read the publication of Bocklandt et al. 2011 to understand the biological provenance of this dataset
- Open RStudio and create a new script called
age.Rfor instance.- Load the
tidyverseandbroompackages withlibrary().
References
Dataset links
- Gene Expression Omnibus accession GSE28746
- Bocklandt S, Lin W, Sehl ME, Sánchez FJ, Sinsheimer JS, Horvath S, Vilain E. (2011) Epigenetic predictor of age. PLoS One. 6(6):e14821. Link
Useful links
- Wikipedia article on Epigenetic Clock
Photo credits
- L’heureuse vieillesse (“happy old age”) by Louis Jean François Lagrenée. Link
Key Points
Several biaises including sequencing depth can result in analysis artifacts and must be corrected trough scaling/normalisation.